Tools and Resources
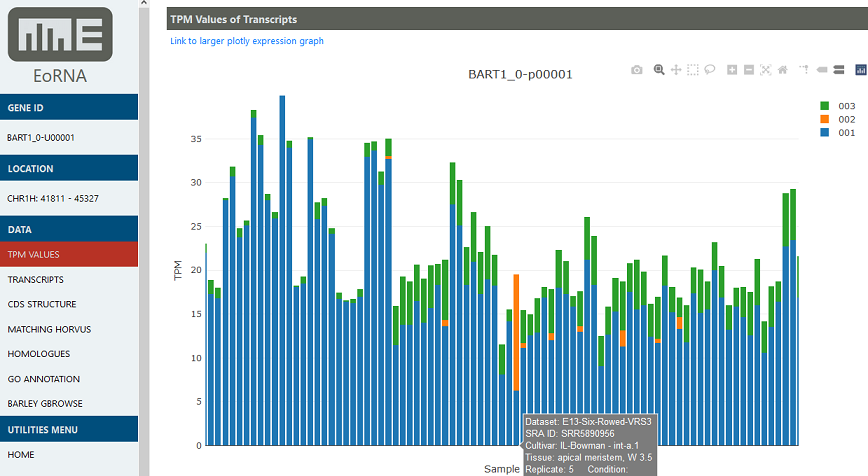
EoRNA is a Barley Expression Database that displays gene and transcript abundance data on demand as transcripts per million (TPM) across all samples and all the genes.
EoRNA provides access to the gene and transcript abundances quantified using data from 22 RNA-seq experiments, covering 843 separate samples, as well as the sequence data and annotation for the Barley Reference Transcript Dataset.
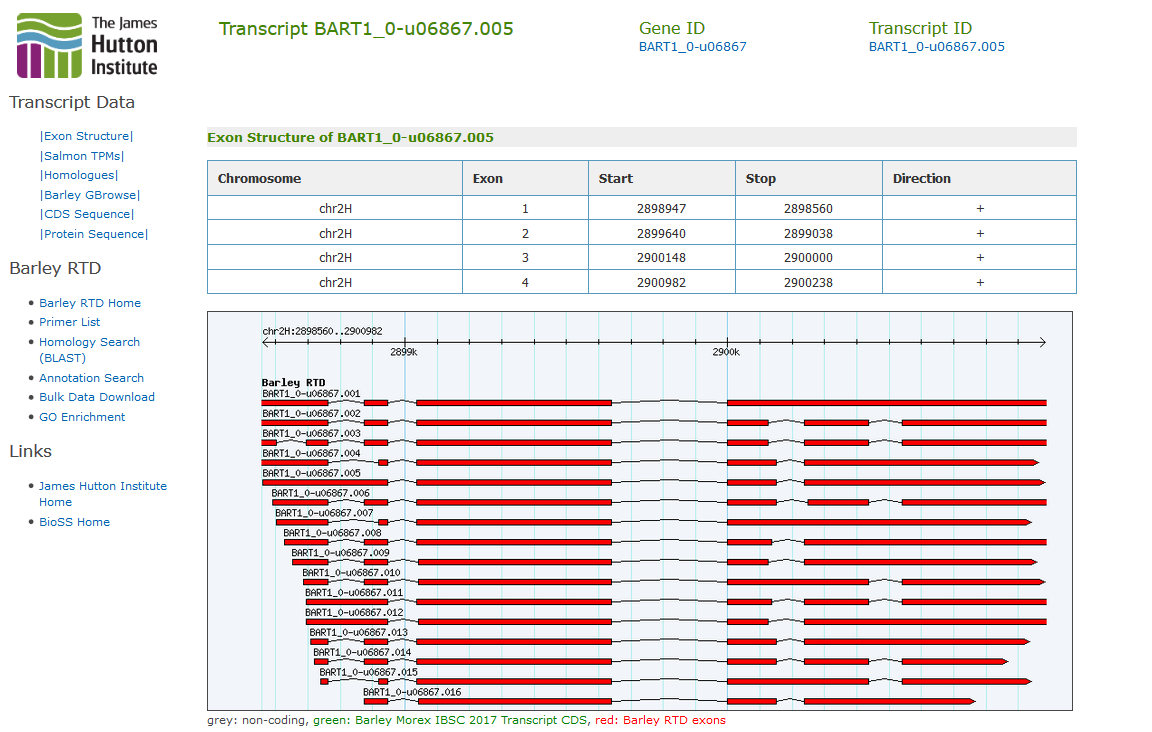
The BarleyRTD website provides access to the Barley Reference Transcript Dataset, containing 177,240 unique barley expressed transcripts covering 60,444 genes. The transcripts are arranged as gene models, and homology to the predicted peptides of Arabidopsis, Rice and Brachypodium is given. Individual transcript expression levels are available as Salmon transcripts per million (TPM) for 16 barley developmental stage samples in Morex.
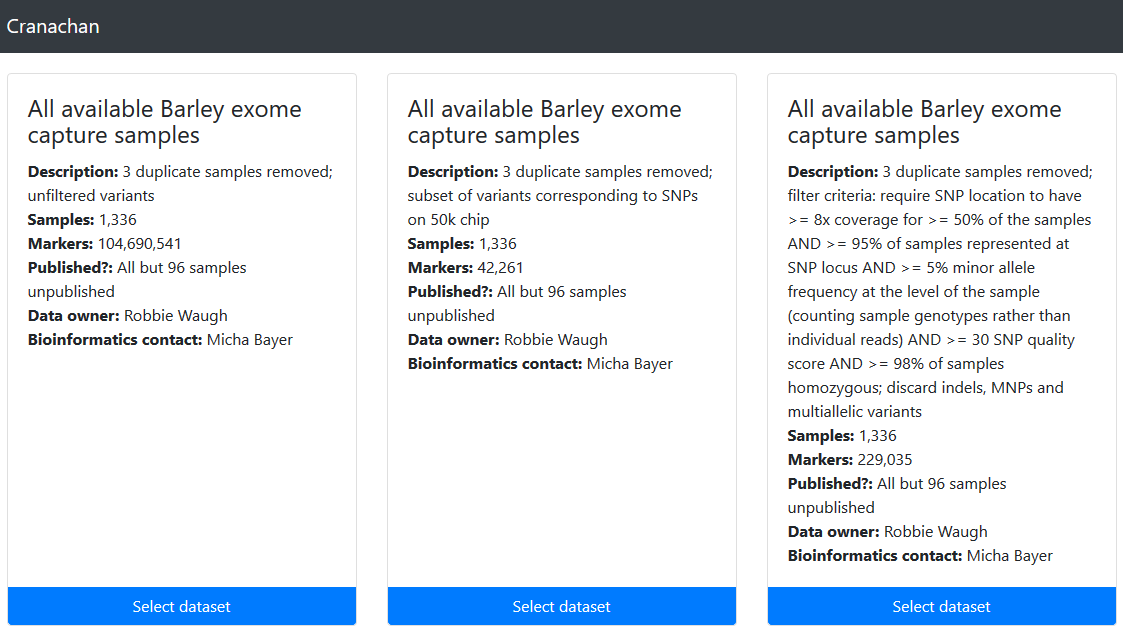
Cranachan provides access to SNPs discovered from the barley exome capture datasets for viewing in Flapjack. Search by gene ID or by genome location.
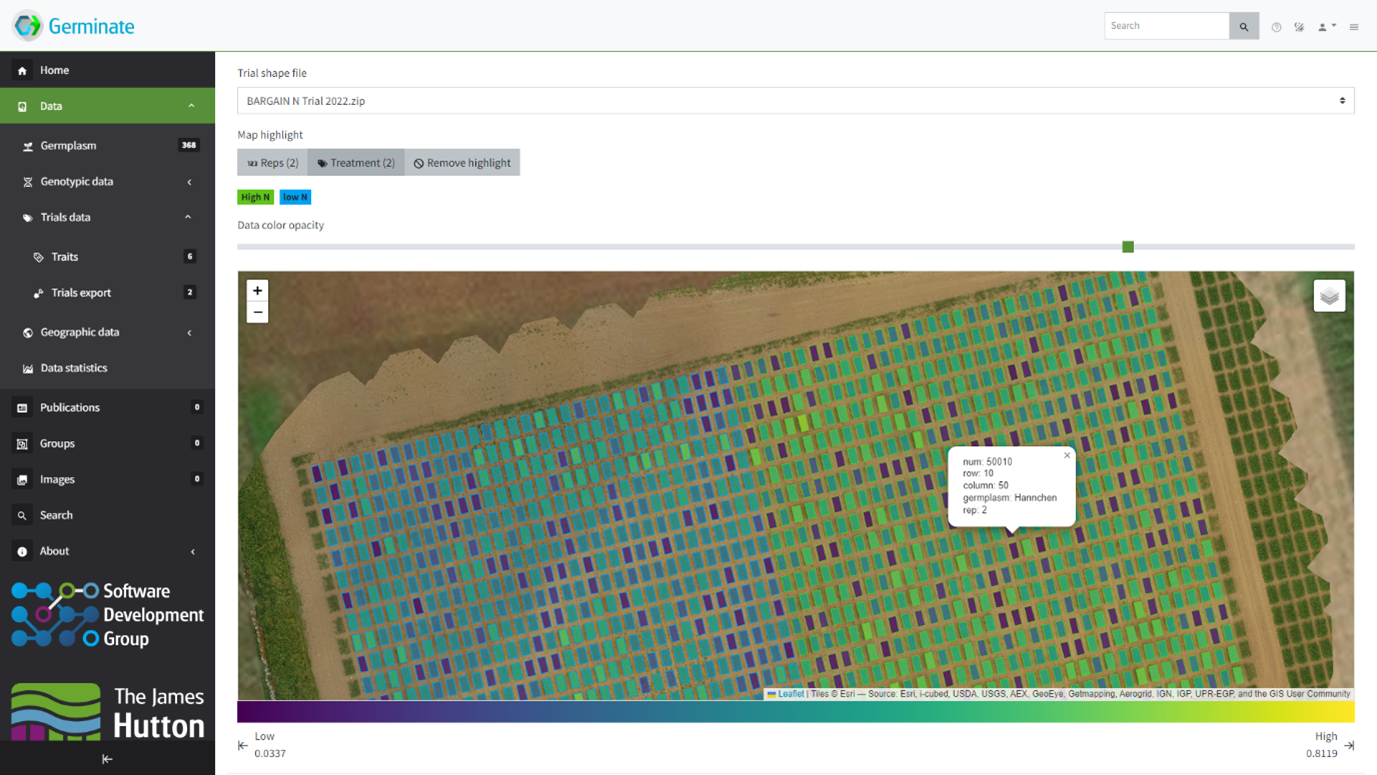
Germinate is a fully featured open-source plant genetic resources platform to store all the data types associated with PGR collections. Examples include (but are not limited to) genotypic, phenotypic, plant passport, environmental, UAV geospatial and time-series type data as well as pedigree and image data. Germinate not only allows the efficient storage and visualization of data but also provides tools to export data in an expanding number of data formats suitable for downstream analysis. Germinate is under constant development and supports several large international plant genetic resources projects.
Access to the 50K SNPs for barley.
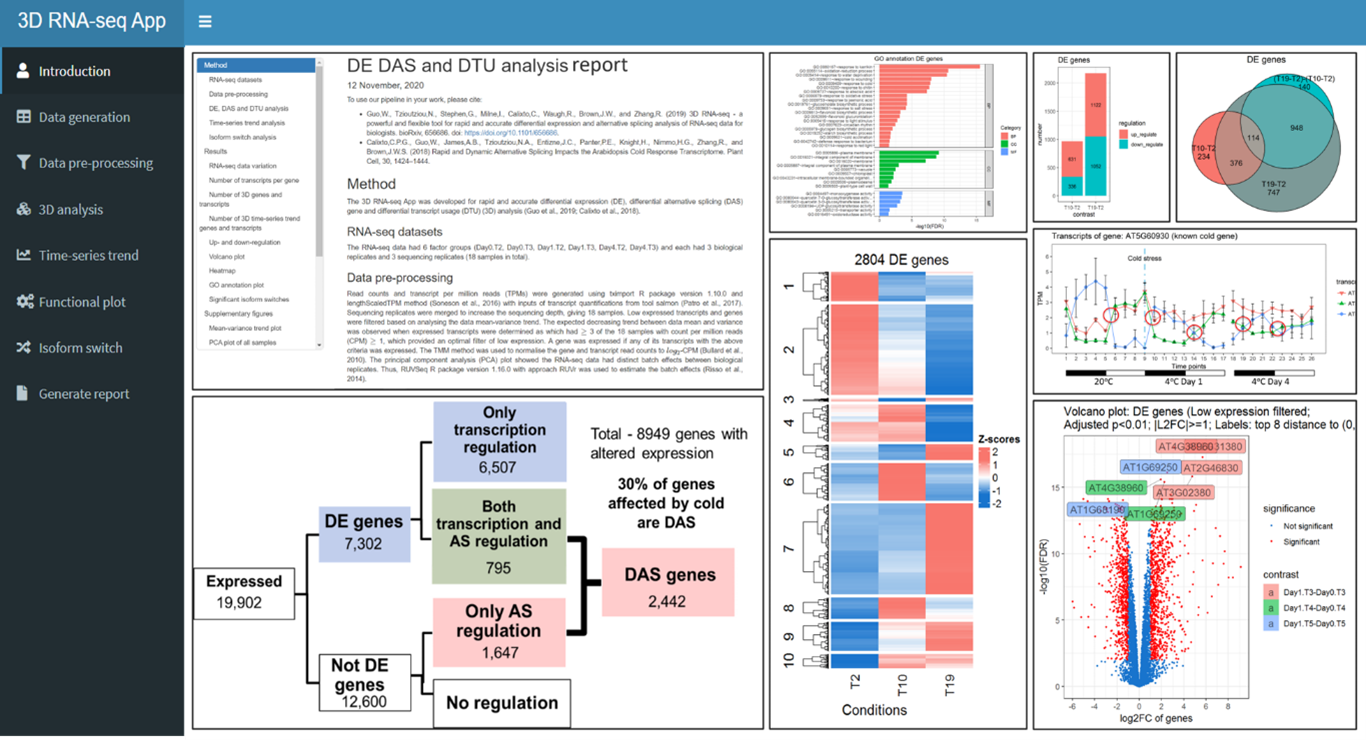
3D RNA-seq is an easy to use, flexible and powerful computational tool for differential gene expression analysis. It integrates the state of art methods for studying gene expression and alternative splicing, making RNA-seq data interpretation simple. Designed for biologists, no specialized skills are required – they can perform professional RNA-seq analysis independently. It accelerates analysis from months to hours, reduces data analysis costs by 80%, and enhances accuracy by over 30%. 3D RNA-seq App has been used beyond barley, aiding rice, soybean, maize, pumpkin, and even human research. 3D RNA-seq has attracted over 15,800 users across 92 countries since 2019 with 10+ training workshops were carried out across the globe.
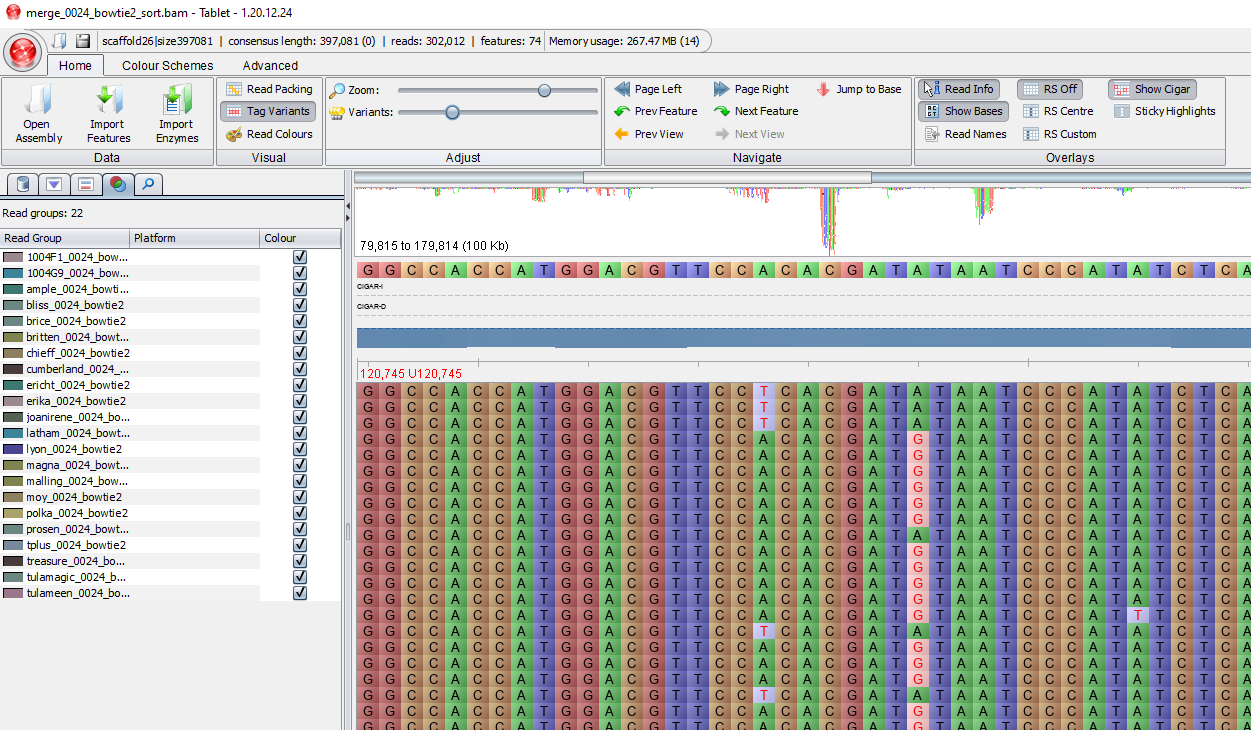
Tablet is a lightweight, high-performance graphical viewer for next generation sequence assemblies and alignments. Good for checking quality of assemblies, read mapping, and polymorphisms. Features whole contig overviews, with data layout and coverage information. Supports a variety of assembly file formats and feature file formats.
GridScore is a new plant phenotyping app utilizing state-of-the-art technologies to improve plant phenotypic data collection. By combining the best features of existing approaches and including a new set of advanced functionalities, GridScore delivers a complete package suitable for manual plant phenotyping experiments.
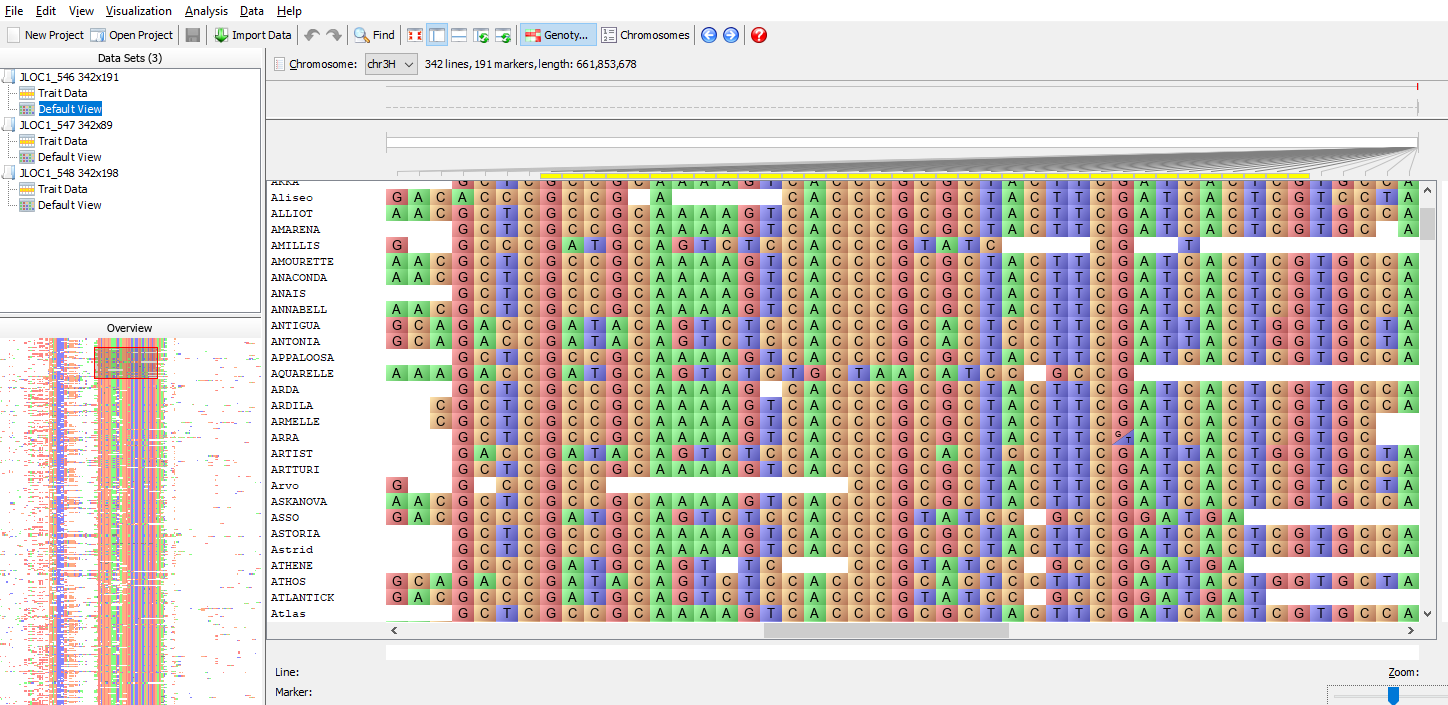
Flapjack is a desktop visualization tool to facilitate analysis of genotype and haplotype data. You can obtain barley genotypic information through the Cranachan Database. Flapjack supports a range of interactions with the data, including graphically moving lines or markers around the display, insertions or deletions of data, and sorting or clustering of lines by either genotype similarity to other lines, or by trait scores. Any map based information such as QTL positions can be aligned against graphical genotypes to identify associated haplotypes. Flapjack also allows for analysis methods to run over the data, such as calculating similarity matrices, running PCoA calculations, or the generation of statistics to help with marker assisted back crossing or pedigree verification of F1s.
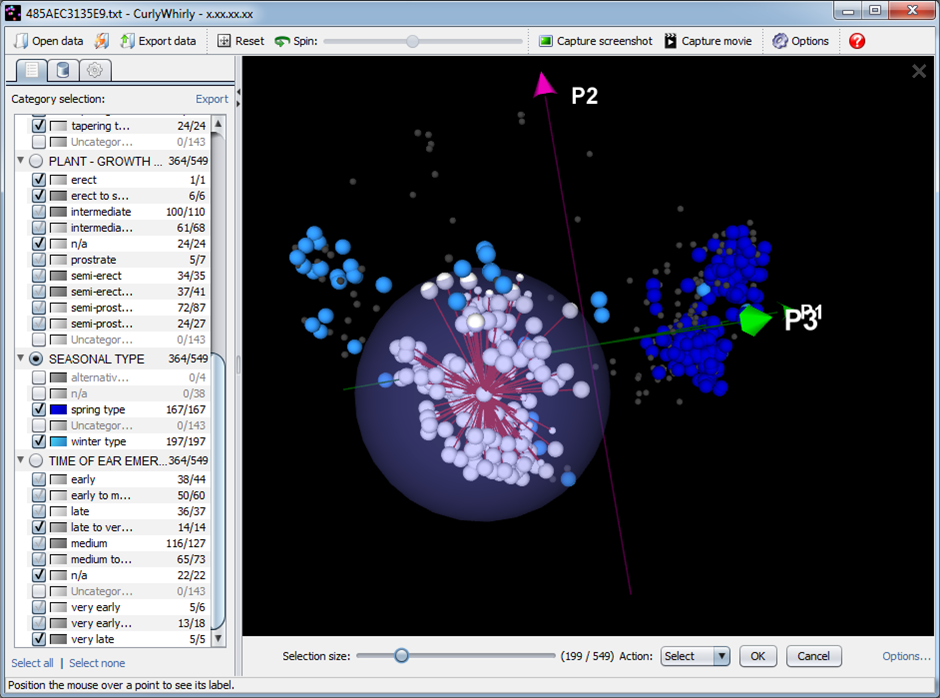
CurlyWhirly is designed for the visualization of 3D data provided as Cartesian coordinate data and is particularly well suited for visualizing the outputs of analyses such as Multidimensional Scaling and Principal Coordinates Analysis. It comfortably handles large-scale genetic diversity data with plots containing tens of thousands of data points on relatively modest hardware.
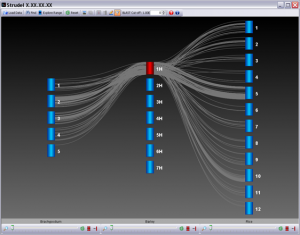
Strudel is our graphical tool for visualizing genetic and physical maps of genomes for comparative purposes. The application aims to let the user examine their data at a variety of different levels of resolution, from entire maps to individual markers, and explore syntenic relationships between genomes.
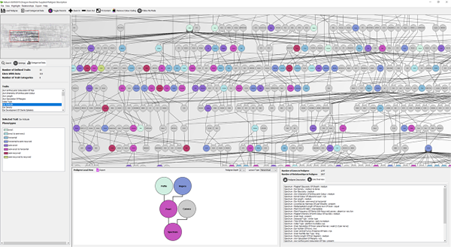
Helium is a generic platform for the visualization of complex plant pedigree data. It offers tools to allow users to explore phenotypic and genotypic information in a pedigree context. Helium allows you to export your pedigree along with user defined colour coding of data. We now have both desktop and web-based versions of Helium available and are working towards introducing new features.
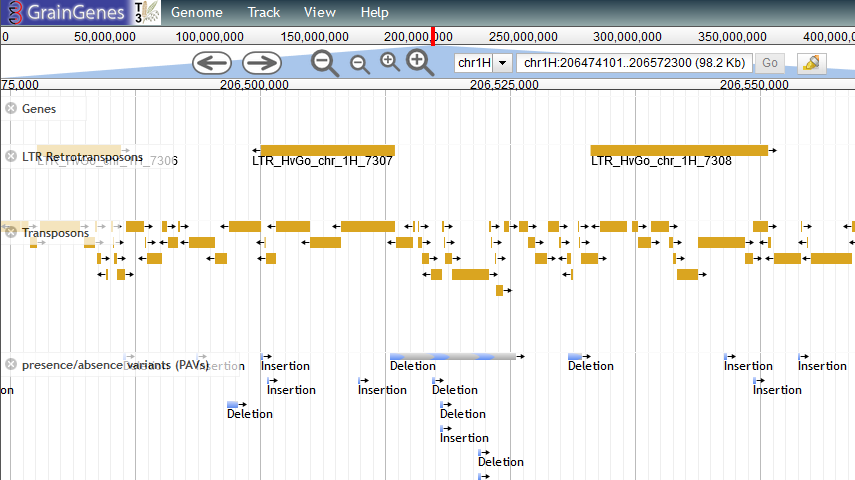
GrainGenes is a centralized repository, and a functional and integrated web interface for wheat, barley, oat, and rye. You can find the 19 genomes here (under "Barley Pangenome Project").
You can reach the BLAST tool for the pan-genomes here: https://wheat.pw.usda.gov/blast
Genesys is an online platform where you can find information about Plant Genetic Resources for Food and Agriculture (PGRFA) conserved in genebanks worldwide. It may be useful for searching for barley germplasm held in genebanks around the world.
The European Search Catalogue for Plant Genetic Resources (EURISCO) provides information about more than 2 million accessions of crop plants and their wild relatives, preserved ex situ by about 400 institutes. It is based on a network of National Inventories of 43 member countries and represents an important effort for the preservation of world's agrobiological diversity by providing information about the large genetic diversity kept by the collaborating institutions.
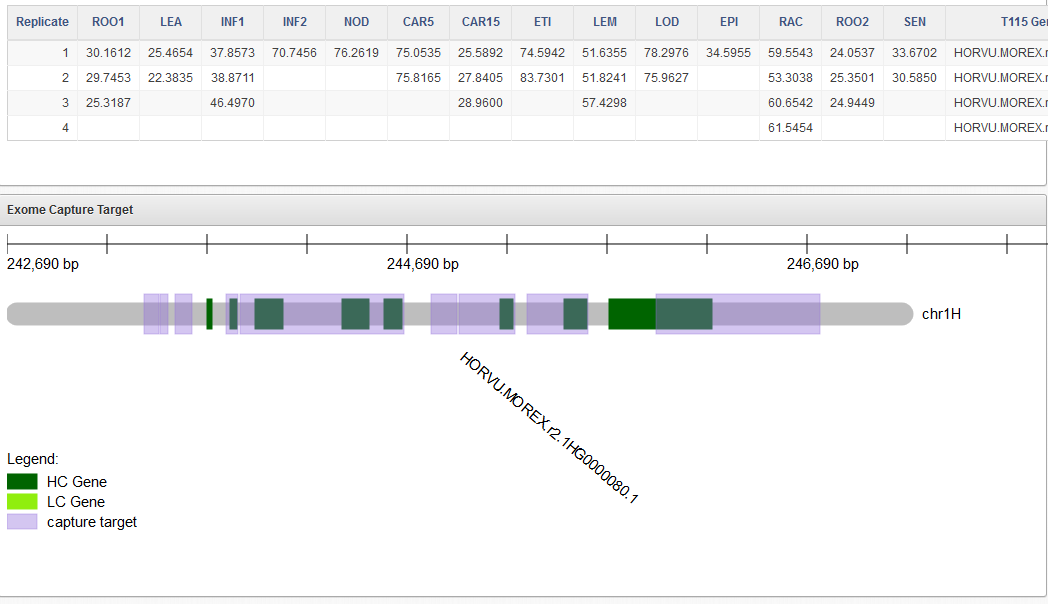
BARLEX (barley genome explorer) is a system to visually inspect the reference genome sequence of barley. BARLEX has been developed by members of the International Barley Sequencing Consortium (IBSC).
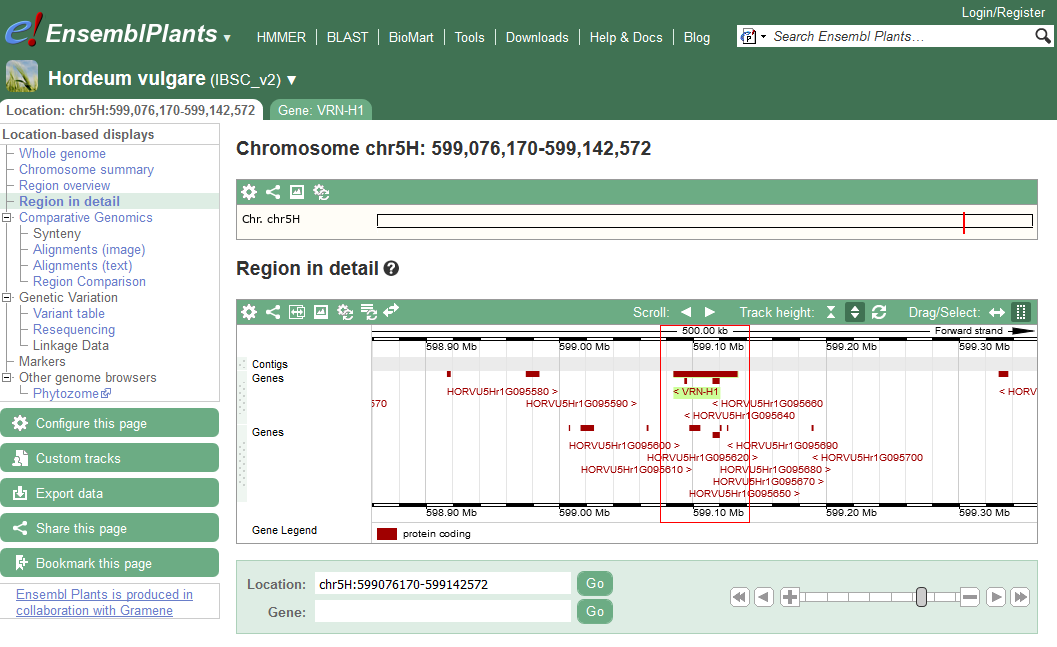
Ensembl is a genome browser for vertebrate genomes that supports research in comparative genomics, evolution, sequence variation and transcriptional regulation. Ensembl annotate genes, computes multiple alignments, predicts regulatory function and collects disease data.
The barley genomic resources currently available in Ensembl Plants are based on the latest gene-space barley assembly from the International Barley Genome Sequencing Consortium.
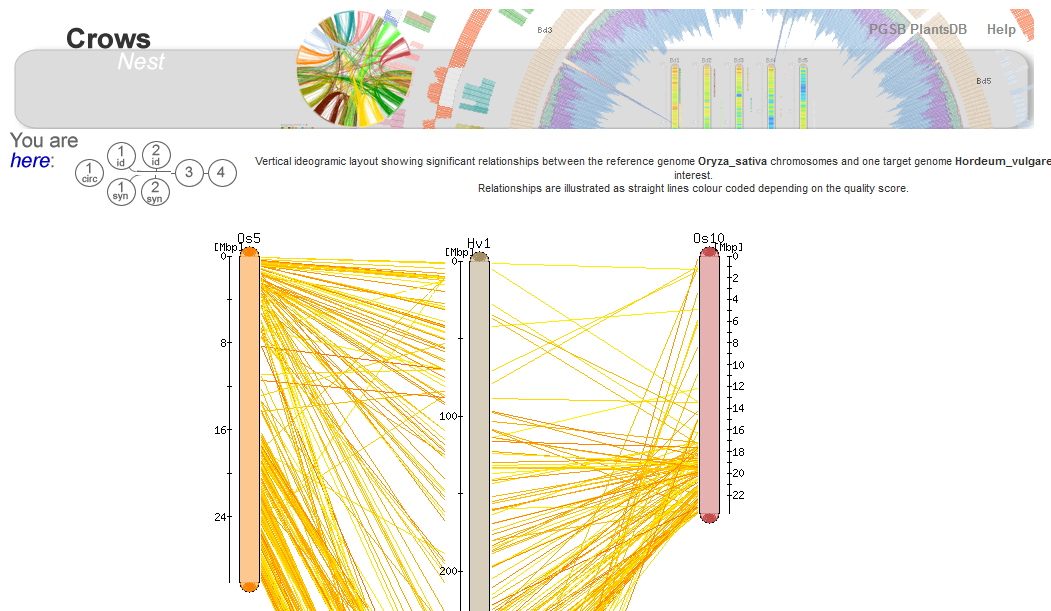
PGSB PlantsDB (http://pgsb.helmholtz-muenchen.de/plant/index.jsp) is a database framework for the comparative analysis and visualization of plant genome data. The resource has been updated with new data sets and types as well as specialized tools and interfaces to address user demands for intuitive access to complex plant genome data.
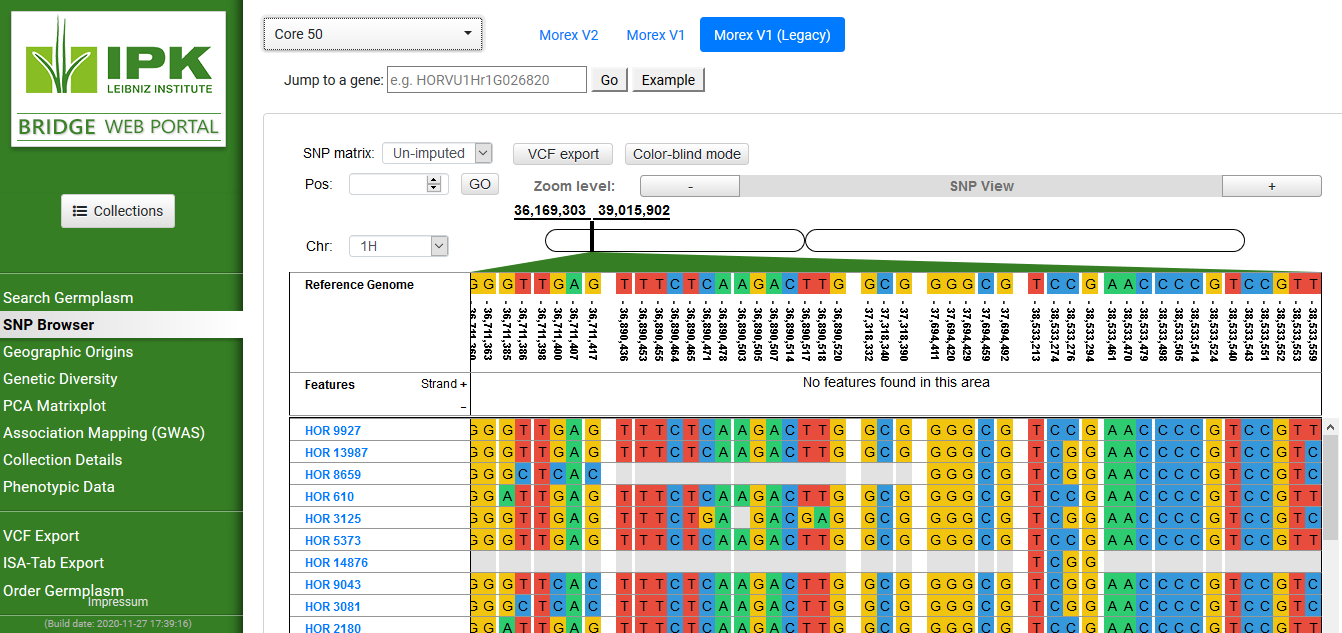
BRIDGE stands for "Biodiversity informatics to bridge the gap from genome information to educated utilization of genetic diversity hosted in Genebanks". It aims at evolving the Federal ex situ Genebank at IPK Gatersleben from a "storage facility" of genetic resources to an integrated resource and information centre, representing a one stop shop for facilitated and informed utilization of crop plant biodiversity.